How to Read Summary Table Anova R
I don't know what fears continue you up at night, merely for me it's worrying that I might accept copy-pasted the wrong values over from my output. No matter how advisedly I check my work, there'due south always the nagging suspicion that I could have confused the contrasts for two different factors, or missed a decimal point or a negative sign.
Although I'm usually overreacting, I recall my paranoia isn't completely misplaced — little errors are much too easy to brand, and they can have horrifying consequences.
Through the years, I've learned that the only sure style to reduce human being mistake is to give humans (including myself) equally piffling opportunity to interfere in the process as possible. Happily, R integrates beautifully with output documents, allowing you to inquire the reckoner to fill in the numbers in your tables and text for y'all, and so you never take to wake up in a cold sweat panicking almost a typo in your correlation matrix. These are called dynamic documents, and they're awesome.
I almost never type out my results anymore; I allow R practice it for me. I wrote my unabridged dissertation in R Studio, in fact, using sweave to integrate my R lawmaking with LaTeX typesetting. I'm writing this post in R Studio as an R-markdown certificate.
Fifty-fifty if you've never used markdown or R-markdown earlier, yous tin bound right in and start getting properly formatted output from R. In this tutorial, I'll comprehend examples for one common model (an analysis of variance, or ANOVA) and testify you how y'all can get tabular array and in-line output automatically.
We'll use one of the most basic functions for creating tables, kable, which is from one of the most convenient packages for combining R code and output together, knitr. (Side note to the fiber enthusiasts: Yes, you're not imagining it — pretty much all of this stuff is playfully named later on yarn, knitting, and material references. Enjoy.) I recommend knitr and kable for people just getting into writing dynamic documents (also chosen literate statistical programming) because they're the easiest and most flexible tools, especially since they tin can exist used to create Discussion documents (not just pdfs or html pages). Depending on your desired output, though, you may discover other packages ameliorate suited to your needs. For case, if you're creating pdf documents, you lot may prefer pander, xtable or stargazer, all of which are much more powerful and elegant. Although these are first-class packages, I find they don't piece of work consistently (or at all) for Word output, which is a bargain breaker for a lot of people.
Quick links to content in this tutorial:
Running an ANOVA in R
Annotated output
Creating an APA style ANOVA tabular array from R output
Inline APA-style output
Recommended further reading
A quick note: I'm using APA style results for the examples hither because that's my groundwork. APA style is also particularly demanding and nit-picky, so befitting to APA standards is a skilful exercise for showing customization options. The code hither can be adapted for pretty much whatsoever formatting you demand, though, so feel gratuitous to take what works for you and leave the rest.
This tutorial assumes…
- That you are using R Studio. If you don't have it already, information technology'south free to downloada and install, just similar R.
- That you already accept a basic understanding of what an ANOVA is and when you might employ information technology.
- That you're not make new to R. If you lot are, the descriptions may still be useful to y'all, merely you may run into problems replicating the assay on your own reckoner or editing the lawmaking to suit your needs.
Running an ANOVA in R
Gear up
Since the kable function is role of the knitr package, you'll demand to load knitr before you can utilise it:
We'll use a data set that comes built into R: warpbreaks. Fittingly, it's about yarn. Information technology gives the number of breaks in yarn tested nether conditions of depression, medium, or high tension, for ii types of wool (A and B). This information comes standard with R, so yous already have it on your estimator. You lot tin read the help documentation about this data set by typing ?warpbreaks in the panel.
str(warpbreaks) # bank check out the structure of the data We'll run a 2x3 factorial ANOVA to test if in that location are differences in the number of breaks based on the type of wool and the amount of tension.
Exploratory data analysis
Before running a model, yous always want to plot the information, to check that your assumptions await okay. Here are a couple plots I might generate while analyzing these data:
library(ggplot2) # histograms, to check out the distribution within each group ggplot(warpbreaks, aes(ten=breaks)) + geom_histogram(bins=x) + facet_grid(wool ~ tension) + theme_classic() 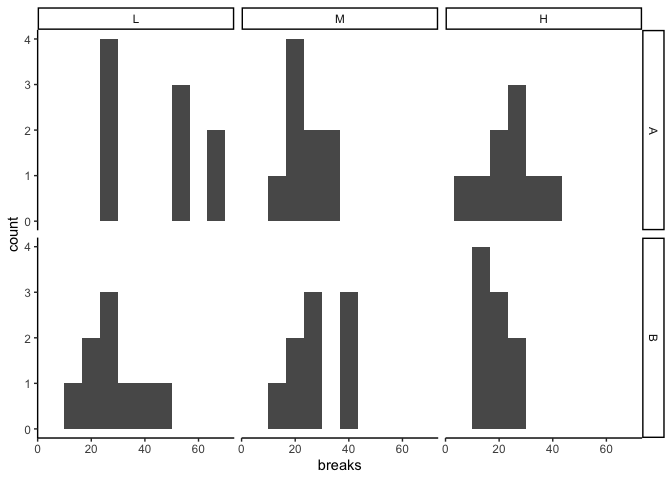
# boxplot, to highlight the group means ggplot(warpbreaks, aes(y=breaks, 10=tension, fill = wool)) + geom_boxplot() + theme_classic() 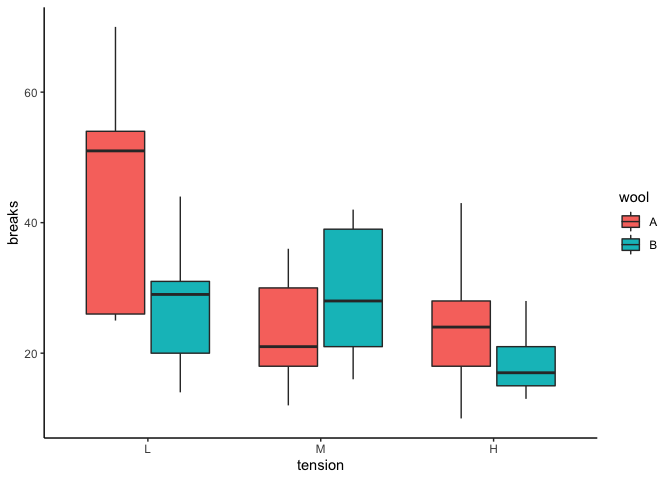
The box plot gives me an idea of what I might find in the ANOVA. It looks like in that location are differences betwixt groups, with fewer breaks at higher tension, and perhaps fewer breaks in wool B vs. wool A at both low and high tension.
The distributions within each cell look pretty wonky, only that'southward non peculiarly surprising given the small-scale sample size (northward=9):
xtabs(~ wool + tension, information = warpbreaks) ## tension ## wool L M H ## A 9 nine 9 ## B ix 9 nine Running the model
One of import consideration when running ANOVAs in R is the coding of factors (in this instance, wool and tension). By default, R uses traditional dummy coding (too called "treatment" coding), which works not bad for regression-style output merely can produce weird sums of squares estimates for ANOVA mode output.
To be on the safe side, always use furnishings coding (contr.sum) or orthogonal dissimilarity coding (e.g.contr.helmert, contr.poly) for factors when running an ANOVA. Hither, I'chiliad choosing to employ effects coding for wool, and polynomial tendency contrasts for tension.
model <- lm ( breaks ~ wool * tension , data = warpbreaks , contrasts = listing ( wool = "contr.sum" , tension = "contr.poly" )) Annotated ANOVA output
The output you'll want to report for an ANOVA depends on the motivation for running the model (is information technology the primary hypothesis exam for your study, or but part of the preliminary descriptive stats?) and the reporting conventions for the journal you intend to submit to. In many cases, you will only want to report the means and standard deviations for each jail cell with notes indicating which master effects and interactions are meaning, and skip reporting the full ANOVA results. Here, I'm going to assume the ANOVA model is primal to your research question, though, and then nosotros can see what a full and detailed study might look like. APA style includes specific guidelines for reporting ANOVA models, which is what I'thou using here.
APA style ANOVA tables generally include the sums of squares, degrees of freedom, F statistic, and p value for each issue. You can become all of those calculations with the Anova function from the automobile package. It'southward important to employ the Anova function rather than the summary.aov office in base R considering Anova allows yous to command the blazon of sums of squares you desire to summate, whereas summary.aov just uses Type 1 (more often than not non what you want, especially if you have an unblanced pattern and/or whatever missing data).
library(motorcar) ## Loading required package: carData sstable <- Anova(model, type = iii) # Type Iii sums of squares is standard, in the social sciences at to the lowest degree sstable ## Anova Table (Blazon III tests) ## ## Response: breaks ## Sum Sq Df F value Pr(>F) ## (Intercept) 42785 1 357.4672 < 2.2e-16 *** ## wool 451 1 3.7653 0.0582130 . ## tension 2034 ii eight.4980 0.0006926 *** ## wool:tension 1003 2 four.1891 0.0210442 * ## Residuals 5745 48 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.one ' ' one The above code runs the Anova function on the model I saved earlier, using Type Three sums of squares, and saves the resulting tabular array equally a new object called sstable.
In sstable, you lot tin encounter a row for each predictor in the model, including the intercept, and the error term (Residuals) at the lesser. The wool:tension term is the interaction betwixt wool and tension (R uses : to specify interaction terms, and * every bit shorthand for an interaction term with both main effects). There are two levels of wool (A and B), so you'll come across ane degree of liberty for that effect. There are 3 levels of tension (low, medium, and loftier), then that has two degrees of freedom. The interaction has the df for both terms multiplied together, i.e. 1 * 2 = 2. The degrees of liberty for the balance are based on the full number of observations in the data (N=54) minus the number of groups, i.eastward. 54-6=48.
The F-statistic for each event is the SSouthward*/*df for that effect divided past the SS*/*df for the residuum. The Pr(>F) gives the p value for that examination, i.east. the probability of observing an F ratio greater than that given the null hypothesis is true.
Contrast estimates
summary.aov(model, split = listing(tension=list(L=1, Q=ii))) ## Df Sum Sq Mean Sq F value Pr(>F) ## wool one 451 450.7 3.765 0.058213 . ## tension 2 2034 1017.ane 8.498 0.000693 *** ## tension: 50 1 1951 1950.vii 16.298 0.000194 *** ## tension: Q 1 84 83.half dozen 0.698 0.407537 ## wool:tension 2 1003 501.4 4.189 0.021044 * ## wool:tension: L 1 251 250.7 two.095 0.154327 ## wool:tension: Q ane 752 752.1 6.284 0.015626 * ## Residuals 48 5745 119.7 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.i ' ' ane (Note that if y'all run summary(model) instead, you'll get the default regression-style output, which is the same information, only represented as regression coefficients with standard errors and t-tests instead of sums of squares estimates with F ratios.)
What we see here is a significant linear tendency in the chief effect for tension — from the box plot we made before, we know that it'southward a negative linear trend. There's no overall quadratic trend in tension, but there is a quadratic trend in the interaction between wool and tension. That means the gauge of the quadratic trend contrast is different for wool A compared to wool B. Referencing the box plot once again, you lot tin meet that the management of the quadratic trend appears to differ in wool A compared to wool B (wool A looks like a positive quadratic trend, with an upward swoop, whereas wool B looks similar a negative quadratic tendency, with an upside-down U dive). There is no linear trend for the interaction, yet, and then that the estimate of the linear tendency in wool A is not significantly different from the estimate of the linear trend in wool B.
Remember that summary.aov is using Type I sums of squares, so the estimates for some effects may not be what we want. In this example, the pattern is balanced and there are no missing data, then the SS estimates using Type I and Blazon III work out to be the same, only in your ain data there may be a deviation. Note that our orthogonal contrasts hither are simple comparisons between ways, and aren't affected past the type of SS used. If you are concerned nearly Type of SS, you may want to grab the dissimilarity estimates from this output and put them into your other sstable object. Here's how you could do that:
Notation which rows in the output correspond to the contrasts you want. In this case, it's rows 3 and 4 for the contrasts on the main outcome of tension, and rows 6 and seven for the contrasts on the interaction. I select those rows with c(3, 4, half dozen, seven). I'm also selecting and reordering the columns in the output, so they'll match what we have in sstable. I select the 2d column (Sum Sq), then the first (Df), then the 4th (F value), then the fifth (Pr(>F)) with c(2, one, 4, 5).
Remember that y'all can employ [ , ] to select item combinations of rows and columns from a given matrix or dataframe. Just put the rows you want every bit the commencement argument, and the columns as the 2nd, i.e.[r, c]. If you leave either the rows or the columns blank, it will render all (so [r, ] will render row r and all columns).
# this pulls out simply the specified rows and columns contrasts <- summary.aov(model, split = list(tension=list(L=1, Q=2)))[[ane]][c(iii, iv, 6, vii), c(2, 1, iv, five)] contrasts ## Sum Sq Df F value Pr(>F) ## tension: Fifty 1950.69 one 16.2979 0.0001938 *** ## tension: Q 83.56 one 0.6982 0.4075366 ## wool:tension: L 250.69 one 2.0945 0.1543266 ## wool:tension: Q 752.08 1 half dozen.2836 0.0156262 * ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' one At present use rbind to create a sort of Frankenstein table, splicing the contrasts estimate rows in with the other rows of sstable.
# select the rows to combine maineffects <- sstable[c(ane,two,three), ] me_contrasts <- contrasts[c(1,two), ] interaction <- sstable[four, ] int_contrasts <- contrasts[c(3,iv), ] resid <- sstable[five, ] # bind the rows together in the desired lodge sstable <- rbind(maineffects, me_contrasts, interaction, int_contrasts, resid) sstable # ta-da! ## Anova Table (Type III tests) ## ## Response: breaks ## Sum Sq Df F value Pr(>F) ## (Intercept) 42785 1 357.4672 < ii.2e-16 *** ## wool 451 1 three.7653 0.0582130 . ## tension 2034 2 8.4980 0.0006926 *** ## tension: L 1951 1 xvi.2979 0.0001938 *** ## tension: Q 84 one 0.6982 0.4075366 ## wool:tension 1003 2 4.1891 0.0210442 * ## wool:tension: L 251 i two.0945 0.1543266 ## wool:tension: Q 752 1 6.2836 0.0156262 * ## Residuals 5745 48 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Estimates of effect size
A popular mensurate of upshot size for ANOVAs (and other linear models) is partial eta-squared. It's the sums of squares for each issue divided by the mistake SS. The post-obit code adds a column to the sstable object with partial eta-squared estimates for each issue:
sstable$pes <- c(sstable$'Sum Sq'[-nrow(sstable)], NA)/(sstable$'Sum Sq' + sstable$'Sum Sq'[nrow(sstable)]) # SS for each outcome divided by the terminal SS (SS_residual) sstable ## Anova Tabular array (Type III tests) ## ## Response: breaks ## Sum Sq Df F value Pr(>F) pes ## (Intercept) 42785 one 357.4672 0.00000 0.88162 ## wool 451 ane 3.7653 0.05821 0.07274 ## tension 2034 2 8.4980 0.00069 0.26149 ## tension: 50 1951 i sixteen.2979 0.00019 0.25348 ## tension: Q 84 i 0.6982 0.40754 0.01434 ## wool:tension 1003 ii 4.1891 0.02104 0.14861 ## wool:tension: Fifty 251 1 2.0945 0.15433 0.04181 ## wool:tension: Q 752 1 6.2836 0.01563 0.11576 ## Residuals 5745 48 Okay great! In that location's your output, simply you lot don't want to simply copy-paste that mess into your manuscript. Let'south get R to generate a overnice, make clean table we can apply in Word.
Creating an APA style ANOVA tabular array from R output
kable(sstable, digits = 3) # the digits argument controls rounding | Sum Sq | Df | F value | Pr(>F) | pes | |
|---|---|---|---|---|---|
| (Intercept) | 42785.185 | ane | 357.467 | 0.000 | 0.882 |
| wool | 450.667 | i | 3.765 | 0.058 | 0.073 |
| tension | 2034.259 | ii | 8.498 | 0.001 | 0.261 |
| tension: L | 1950.694 | ane | xvi.298 | 0.000 | 0.253 |
| tension: Q | 83.565 | 1 | 0.698 | 0.408 | 0.014 |
| wool:tension | 1002.778 | two | 4.189 | 0.021 | 0.149 |
| wool:tension: L | 250.694 | 1 | 2.095 | 0.154 | 0.042 |
| wool:tension: Q | 752.083 | 1 | half-dozen.284 | 0.016 | 0.116 |
| Residuals | 5745.111 | 48 | NA | NA | NA |
Wait, what? That was and so like shooting fish in a barrel!
Yes, yes it was. :)
In a lot of cases, that will exist all you need to become a workable ANOVA table in your certificate. Merely for fun, though, let's play effectually with customizing it a trivial.
Hibernate missing values
By default, kable displays missing values in a table every bit NA, but in this case we'd rather accept them just be blank. You can control that with the options command:
options(knitr.kable.NA = '') # this volition hibernate missing values in the kable table kable(sstable, digits = 3) | Sum Sq | Df | F value | Pr(>F) | foot | |
|---|---|---|---|---|---|
| (Intercept) | 42785.185 | 1 | 357.467 | 0.000 | 0.882 |
| wool | 450.667 | one | 3.765 | 0.058 | 0.073 |
| tension | 2034.259 | 2 | eight.498 | 0.001 | 0.261 |
| tension: Fifty | 1950.694 | ane | sixteen.298 | 0.000 | 0.253 |
| tension: Q | 83.565 | i | 0.698 | 0.408 | 0.014 |
| wool:tension | 1002.778 | 2 | 4.189 | 0.021 | 0.149 |
| wool:tension: L | 250.694 | 1 | ii.095 | 0.154 | 0.042 |
| wool:tension: Q | 752.083 | 1 | 6.284 | 0.016 | 0.116 |
| Residuals | 5745.111 | 48 |
Add a tabular array caption
You tin add a title for the table if you alter the format to "pandoc". Depending on your final document output (pdf, html, Word, etc.), y'all can get automatic table numbering this way as well, which saves much time and many headaches.
kable(sstable, digits = iii, format = "pandoc", explanation = "ANOVA table") | Sum Sq | Df | F value | Pr(>F) | pes | |
|---|---|---|---|---|---|
| (Intercept) | 42785.185 | 1 | 357.467 | 0.000 | 0.882 |
| wool | 450.667 | 1 | 3.765 | 0.058 | 0.073 |
| tension | 2034.259 | 2 | eight.498 | 0.001 | 0.261 |
| tension: L | 1950.694 | 1 | 16.298 | 0.000 | 0.253 |
| tension: Q | 83.565 | 1 | 0.698 | 0.408 | 0.014 |
| wool:tension | 1002.778 | 2 | iv.189 | 0.021 | 0.149 |
| wool:tension: L | 250.694 | i | 2.095 | 0.154 | 0.042 |
| wool:tension: Q | 752.083 | ane | 6.284 | 0.016 | 0.116 |
| Residuals | 5745.111 | 48 |
ANOVA table
Alter cavalcade and row names
Often, the automated row names and column names aren't quite what yous want. If so, you'll demand to change them for the sstable object itself, and and so run kable on the updated object.
colnames(sstable) <- c("SS", "df", "$F$", "$p$", "fractional $\\eta^2$") rownames(sstable) <- c("(Intercept)", "Wool", "Tension", "Tension: Linear Tendency", "Tension: Quadratic Trend", "Wool 10 Tension", "Wool x Tension: Linear Tendency", "Wool x Tension: Quadratic Tendency", "Residuals") kable(sstable, digits = iii, format = "pandoc", caption = "ANOVA table") | SS | df | F | p | fractional η ii | |
|---|---|---|---|---|---|
| (Intercept) | 42785.185 | ane | 357.467 | 0.000 | 0.882 |
| Wool | 450.667 | ane | 3.765 | 0.058 | 0.073 |
| Tension | 2034.259 | 2 | viii.498 | 0.001 | 0.261 |
| Tension: Linear Trend | 1950.694 | 1 | 16.298 | 0.000 | 0.253 |
| Tension: Quadratic Trend | 83.565 | one | 0.698 | 0.408 | 0.014 |
| Wool 10 Tension | 1002.778 | 2 | 4.189 | 0.021 | 0.149 |
| Wool x Tension: Linear Trend | 250.694 | one | 2.095 | 0.154 | 0.042 |
| Wool 10 Tension: Quadratic Trend | 752.083 | ane | 6.284 | 0.016 | 0.116 |
| Residuals | 5745.111 | 48 |
ANOVA table
Omit the intercept row
For many models, the intercept is not of any theoretical involvement, and you may want to omit it from the output. If you just want to driblet one row (or column), the easiest arroyo is to signal that row'due south number and put a minus sign before it:
kable(sstable[-one, ], digits = 3, format = "pandoc", caption = "ANOVA table") | SS | df | F | p | partial η 2 | |
|---|---|---|---|---|---|
| Wool | 450.667 | 1 | 3.765 | 0.058 | 0.073 |
| Tension | 2034.259 | two | 8.498 | 0.001 | 0.261 |
| Tension: Linear Trend | 1950.694 | i | 16.298 | 0.000 | 0.253 |
| Tension: Quadratic Tendency | 83.565 | one | 0.698 | 0.408 | 0.014 |
| Wool x Tension | 1002.778 | 2 | 4.189 | 0.021 | 0.149 |
| Wool x Tension: Linear Trend | 250.694 | 1 | 2.095 | 0.154 | 0.042 |
| Wool x Tension: Quadratic Tendency | 752.083 | one | 6.284 | 0.016 | 0.116 |
| Residuals | 5745.111 | 48 |
ANOVA table
Inline APA-fashion output
You tin can also knit R output right into your typed sentences! To include inline R output, use back-ticks, like this:
Here'due south a sentence, and I want to allow you know that the total number of cats I own is `r length(my_cats)`. The back-ticks marker out the code to run, and the r after the start back-tick tells knitr that it's R lawmaking (if y'all feel the demand, you can incorporate code from pretty much any language yous like, not just R). Assuming there'south a vector called my_cats, when nosotros knit the document, that line of code will be evaluated and the result (the number of items in the vector my_cats) will be printed right in that sentence.
Let'south work this into our ANOVA reporting.
Here's an instance write-up of this ANOVA, using inline code to plug in the stats. Since the inline lawmaking can be a little hard to read, I similar to save all of the variables I want to use inline with convenient names first.
fstat <- unname(summary(model)$fstatistic[one]) df_model <- unname(summary(model)$fstatistic[2]) df_res <- unname(summary(model)$fstatistic[3]) rsq <- summary(model)$r.squared p <- pf(fstat, df_model, df_res, lower.tail = Imitation) Then I can plug them in as needed in my writing.
A 2x3 factorial ANOVA revealed that wool type (A or B) and tension (depression, medium, or high) predict a significant corporeality of variance in number of breaks, $R^two$=`r circular(rsq, 2)`, $F(`r round(df_model, 0)`, `r round(df_res, 0)`)=`r round(fstat, 2)`$, $p=`r ifelse(round(p, iii) == 0, "<.001", round(p, iii))`$. Hither'southward how that lawmaking renders when knit: A 2x3 factorial ANOVA revealed that wool type (A or B) and tension (low, medium, or high) predict a significant amount of variance in number of breaks, R two=0.38, F(5, 48) = 5.83, p = < .001.
Further Reading
You can use this strategy just to generate tables or petty bits of output and and so copy-paste them into your manuscript draft in Word (or wherever you write), and that will get a long way toward reducing the possibility for man error in your scientific writing. That's a swell way to go started, and if your own work habits (or those of your co-authors) rely strongly on MS tools that may be as far as you have it.
Once you outset doing that, though, you'll realize you still get caught in a trap where you have to remember to update all the output in your document every time you make a change in the data or assay (e.g. your PI asks you to remove an outlier and re-run everything, so you lot painstakingly re-practice every piece of output, then for the side by side typhoon she asks yous to put the outlier back in and you have to check it all again). The power of literate statistical programming and dynamic documents really comes through when yous can write your whole document in the aforementioned environment yous practice your assay (e.g. RStudio). And then, when you accept an update, you can simply change the one line of code at the peak where, for example, you exclude that outlier or not, and when you "knit" the certificate all of the output will automatically update to reverberate the change. It takes minutes instead of hours, and there's no re-create-pasting for you to accidentally mess upwards. Dissimilar with Discussion documents, R-markdown documents are obviously text and so they requite you the option to accept advantage of version command tools like git to keep rigorous records of all the changes made to both your assay and your writing. Your entire change history is bachelor to you in a tidy, transparent way, and you lot can even develop parallel versions of a document and then merge them down the road and never lose the history.
At that place are tons of slap-up tools to assistance you write a huge range of content in R-markdown. Here are some specific references for popular formats and extensions:
- scientific manuscripts, including lots of ready templates that will match the formatting requirements for specific journals
- math expressions and equations
- citations and references, including automatic bibliographies
- presentation slides
- books, including like shooting fish in a barrel application for ePub and Kindle formats
- blogs and websites
- online tutorials, including the selection for interactive feedback
For more on creating tables using kable, every bit nosotros did here, run across this post. For more than on inline R code, see this postal service. For a great general reference, see the RStudio cheatsheet for R-markdown. Note that if you open RStudio, you tin can access all of their excellent cheatsheets correct at that place from the card at the meridian: become to Help, then select Cheatsheets.
Source: https://education.arcus.chop.edu/anova-tables-in-r/
Belum ada Komentar untuk "How to Read Summary Table Anova R"
Posting Komentar